CNN for FCAL Shower Classification#
Prepare Dataset#
%pip install h5py scikit-learn torch torchvision tqdm safetensors > /dev/null
Note: you may need to restart the kernel to use updated packages.
import os
from urllib.parse import urlparse
import urllib.request
from tqdm import tqdm
class DownloadProgressBar(tqdm):
"""Custom TQDM progress bar for urllib downloads."""
def update_to(self, blocks=1, block_size=1, total_size=None):
"""
Update the progress bar.
Args:
blocks (int): Number of blocks transferred so far.
block_size (int): Size of each block (in bytes).
total_size (int, optional): Total size of the file (in bytes).
"""
if total_size is not None:
self.total = total_size
self.update(blocks * block_size - self.n)
def download(url, target_dir):
"""
Download a file from a URL into the target directory with progress display.
Args:
url (str): Direct URL to the file.
target_dir (str): Directory to save the file.
Returns:
str: Path to the downloaded (or existing) file.
"""
# Ensure the target directory exists
os.makedirs(target_dir, exist_ok=True)
# Infer the filename from the URL
filename = os.path.basename(urlparse(url).path)
local_path = os.path.join(target_dir, filename)
# If file already exists, skip download
if os.path.exists(local_path):
print(f"✅ File already exists: {local_path}")
return local_path
# Download with progress bar
print(f"⬇️ Downloading {filename} from {url}")
with DownloadProgressBar(unit='B', unit_scale=True, miniters=1, desc=filename) as t:
urllib.request.urlretrieve(url, filename=local_path, reporthook=t.update_to)
print(f"✅ Download complete: {local_path}")
return local_path
dataset_url = "https://huggingface.co/datasets/AI4EIC/DNP2025-tutorial/resolve/main/formatted_dataset/CNN4FCAL_GUN_PATCHSIZE_11.h5"
data_dir = "data"
dataset_path = download(dataset_url, data_dir)
⬇️ Downloading CNN4FCAL_GUN_PATCHSIZE_11.h5 from https://huggingface.co/datasets/AI4EIC/DNP2025-tutorial/resolve/main/formatted_dataset/CNN4FCAL_GUN_PATCHSIZE_11.h5
CNN4FCAL_GUN_PATCHSIZE_11.h5: 30.3MB [00:00, 37.1MB/s]
✅ Download complete: data/CNN4FCAL_GUN_PATCHSIZE_11.h5
Step 1: Open and Inspect the .h5 File#
We can open the dataset and confirm whats inside it
import h5py, numpy as np
with h5py.File(dataset_path, "r") as f:
print("✅ Datasets available:", list(f.keys()))
print("🔍 patches shape:", f["patches"].shape)
print("🔍 labels shape:", f["label"].shape)
✅ Datasets available: ['label', 'patches', 'showerE', 'thrownE']
🔍 patches shape: (814151, 11, 11)
🔍 labels shape: (814151,)
Step 2: Create a Custom PyTorch Dataset#
This dataset will:
Read directly from the
.h5fileOptionally we can apply normalization in the form of transformation
import torch
from torch.utils.data import Dataset, DataLoader
import numpy as np # Import numpy
class FCALPatchDataset(Dataset):
def __init__(self, h5_path, indices=None, transform=None):
"""
Parameters
----------
h5_path : str
Path to the HDF5 file.
indices : list or np.ndarray, optional
Subset of indices to use (for train/val/test split).
transform : callable, optional
Optional transform to apply to each patch.
"""
# Open the file handle here and keep it open
self.h5_path = h5_path
self.file = h5py.File(self.h5_path, "r")
self.indices = indices
self.transform = transform
# Assign datasets directly from the file handle
self.length = len(self.file["label"])
self.patches = np.array(self.file["patches"], dtype=np.float32)
self.patches = self.patches[self.indices] if self.indices is not None else self.patches
self.labels = np.array(self.file["label"], dtype=np.int64)
self.labels = self.labels[self.indices if self.indices is not None else slice(None)]
self.showerE = np.array(self.file["showerE"], dtype=np.float32)
self.showerE = self.showerE[self.indices if self.indices is not None else slice(None)]
self.thrownE = np.array(self.file["thrownE"], dtype=np.float32)
self.thrownE = self.thrownE[self.indices if self.indices is not None else slice(None)]
self.file.close()
def __len__(self):
return self.length
def __getitem__(self, idx):
# Handle subset indices
# Access data directly from the assigned attributes
patch = self.patches[idx]
label = self.labels[idx]
showerE = self.showerE[idx]
thrownE = self.thrownE[idx]
# Optional transform
if self.transform:
patch = self.transform(patch)
# Convert to torch tensors
patch = torch.from_numpy(patch).unsqueeze(0) # (1, 11, 11)
return patch, label, showerE, thrownE
Step 3: Split into Train / Validation / Testing#
from sklearn.model_selection import train_test_split
import numpy as np
# Read total number of samples
with h5py.File(dataset_path, "r") as f:
N = len(f["label"])
labels = np.array(f["label"]) # ✅ Read dataset into memory
indices = np.arange(N)
n_photons = np.sum(labels == 1)
n_splitOffs = np.sum(labels == 0)
counts = np.array([n_splitOffs, n_photons], dtype = np.float32)
# 70% train, 15% val, 15% test (stratified by class labels)
train_idx, temp_idx = train_test_split(
indices, test_size=0.3, stratify=labels, random_state=42
)
val_idx, test_idx = train_test_split(
temp_idx, test_size=0.5, stratify=labels[temp_idx], random_state=42
)
Step 4: Create Dataset and DataLoader Objects#
Let us normalize each of the images with the total energy of all the hits in the shower
ENERGY_SCALE = 0.05 # GeV (for global log scaling)
CLIP_MAX = 2.0 # GeV per cell clamp for scaling
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
USE_AMP = (DEVICE == "cuda") # automatic mixed precision only on CUDA
BATCH_SIZE = 512
NUM_WORKERS = 1
pin = (DEVICE == "cuda")
def log_global_norm(W, e0=ENERGY_SCALE, emax=CLIP_MAX):
# Keep everything float32 to avoid silent upcasts to float64
W = W.clip(min=0).astype(np.float32, copy=False)
e0 = np.float32(e0)
emax = np.float32(emax)
Z = np.log1p(W / e0).astype(np.float32, copy=False)
Z /= np.log1p(emax / e0).astype(np.float32, copy=False)
np.clip(Z, 0.0, 1.0, out=Z)
return Z # float32
def normalize_patch(patch):
# Example normalization to total energy = 1
total = np.sum(patch)
return patch / total if total > 0 else patch
train_ds = FCALPatchDataset(dataset_path, indices=train_idx, transform=log_global_norm)
val_ds = FCALPatchDataset(dataset_path, indices=val_idx, transform=log_global_norm)
test_ds = FCALPatchDataset(dataset_path, indices=test_idx, transform=log_global_norm)
train_loader = DataLoader(train_ds, batch_size=BATCH_SIZE, shuffle=True
)
val_loader = DataLoader(val_ds, batch_size=BATCH_SIZE, shuffle=False
)
test_loader = DataLoader(test_ds, batch_size=BATCH_SIZE, shuffle=False
)
# lets check a few samples
patches, labels, showerE, thrownE = next(iter(train_loader))
print("Batch shape:", patches.shape) # (B, 1, 11, 11)
print("Labels:", labels[:8])
print("showerE:", showerE[:8])
print("thrownE:", thrownE[:8])
Batch shape: torch.Size([512, 1, 11, 11])
Labels: tensor([0, 1, 1, 1, 1, 1, 1, 1])
showerE: tensor([0.1467, 1.1496, 0.9625, 0.3645, 1.5525, 1.4257, 1.0802, 1.8474])
thrownE: tensor([1.6663, 1.1155, 1.1404, 0.9574, 1.5913, 1.4105, 1.1804, 1.8689])
CNN Model Architecture and training#
The architerure will have the following architecture
Layer Type |
Output Channels |
Kernel / Operation |
Purpose |
|---|---|---|---|
|
16 |
3×3 |
Extract local energy patterns and normalize activations. |
|
— |
2×2 downsample |
Reduce spatial resolution and emphasize dominant features. |
|
32 |
3×3 |
Capture higher-level correlations (e.g., multi-peak structure). |
|
— |
— |
Further compress spatial information. |
|
64 |
3×3 |
Learn more abstract, class-discriminating representations. |
|
— |
Global average |
Collapse the spatial dimension to a single latent vector (size 64). |
|
— |
Fully connected |
Classify into photon (1) or split-off (0). |
import torch, torch.nn as nn
# ---------------------------
# Model
# ---------------------------
class SmallCNN(nn.Module):
def __init__(self, in_ch=1, n_classes=2):
super().__init__()
self.net = nn.Sequential(
nn.Conv2d(in_ch, 16, 3, padding=1), nn.BatchNorm2d(16), nn.ReLU(inplace=True),
nn.MaxPool2d(2),
nn.Conv2d(16, 32, 3, padding=1), nn.BatchNorm2d(32), nn.ReLU(inplace=True),
nn.MaxPool2d(2),
nn.Conv2d(32, 64, 3, padding=1), nn.BatchNorm2d(64), nn.ReLU(inplace=True),
nn.AdaptiveAvgPool2d(1)
)
self.head = nn.Linear(64, n_classes)
def forward(self, x):
return self.head(self.net(x).flatten(1))
from torch.nn.functional import one_hot as one_hot
from tqdm.auto import tqdm, trange
from sklearn.metrics import roc_auc_score, confusion_matrix
from contextlib import nullcontext
import math
from safetensors.torch import save_model
# ---------------------------
# Eval & Train (with progress bars + new AMP)
# ---------------------------
EPOCHS = 20
LR = 3e-4
DO_TRAIN = True
@torch.inference_mode()
def evaluate(model, loader, desc="Eval", returnShowerE = False, returnthrownE = False):
model.eval()
model.to(DEVICE)
ys, ps = [], []
correct = total = 0
bar = tqdm(loader, desc=desc, leave=False)
shE = []
thE = []
for Xb, yb, showerE, thrownE in bar:
Xb = Xb.to(DEVICE, non_blocking=True, dtype=torch.float32)
yb = yb.to(DEVICE, non_blocking=True)
logits = model(Xb)
prob = torch.softmax(logits, dim=1)[:, 1]
pred = logits.argmax(dim=1)
ys.append(yb.cpu().numpy()); ps.append(prob.cpu().numpy())
shE.append(showerE.cpu().numpy())
thE.append(thrownE.cpu().numpy())
correct += (pred == yb).sum().item(); total += yb.numel()
bar.set_postfix(acc=f"{(correct/max(1,total)):.3f}")
y_true = np.concatenate(ys) if ys else np.array([])
y_prob = np.concatenate(ps) if ps else np.array([])
showerE = np.concatenate(shE) if shE else np.array([])
thrownE = np.concatenate(thE) if thE else np.array([])
acc = correct / max(1, total)
auc = roc_auc_score(y_true, y_prob) if y_true.size and np.unique(y_true).size > 1 else float("nan")
if returnShowerE:
return acc, auc, y_true, y_prob, showerE
elif returnthrownE:
return acc, auc, y_true, y_prob, thrownE
else:
return acc, auc, y_true, y_prob
def train(model, opt, train_loader, val_loader, save_path = "./models", counts = counts, epochs = EPOCHS):
os.makedirs(save_path, exist_ok=True)
weights = torch.tensor((counts.sum() / (counts + 1e-6)), dtype=torch.float32, device=DEVICE)
crit = nn.CrossEntropyLoss(weight=weights)
scaler = torch.amp.GradScaler('cuda') if USE_AMP else None
model.to(DEVICE)
best_auc, best_state, patience, bad = -1.0, None, 5, 0
for epoch in trange(1, epochs+1, desc="Training"):
model.train()
running, seen, correct = 0.0, 0, 0
bar = tqdm(train_loader, desc=f"Epoch {epoch}/{EPOCHS} (train)", leave=False)
for Xb, yb, _, _ in bar:
Xb = Xb.to(DEVICE, non_blocking=True, dtype=torch.float32)
yb = yb.to(DEVICE, non_blocking=True)
opt.zero_grad(set_to_none=True)
ctx = torch.amp.autocast(device_type='cuda', dtype=torch.float16) if USE_AMP else nullcontext()
with ctx:
logits = model(Xb)
loss = crit(logits, yb)
if USE_AMP:
scaler.scale(loss).backward()
scaler.step(opt)
scaler.update()
else:
loss.backward()
opt.step()
running += loss.item() * yb.size(0)
seen += yb.size(0)
correct += (logits.argmax(1) == yb).sum().item()
bar.set_postfix(avg_loss=f"{running/max(1,seen):.4f}", acc=f"{(correct/max(1,seen)):.3f}")
train_loss = running / max(1, seen)
val_acc, val_auc, _, _ = evaluate(model, val_loader, desc=f"Epoch {epoch}/{EPOCHS} (val)")
tqdm.write(f"Epoch {epoch:02d} | train_loss {train_loss:.4f} | val_acc {val_acc:.4f} | val_auc {val_auc:.4f}")
score = 0.0 if math.isnan(val_auc) else val_auc
if score > best_auc:
best_auc, bad = score, 0
best_state = {k: v.detach().cpu().clone() for k,v in model.state_dict().items()}
tqdm.write(f"✓ New best AUC {best_auc:.4f}")
save_model(model, os.path.join(save_path, "FCALClassifier.safetensors"))
else:
bad += 1
if bad >= patience:
tqdm.write("Early stopping.")
break
if best_state is not None:
model.load_state_dict(best_state)
return model
model = SmallCNN().to(DEVICE).to(torch.float32)
opt = torch.optim.AdamW(model.parameters(), lr=LR)
save_path = "/home/ksuresh/data10/DNP2025/models"
if DO_TRAIN:
model = train(model, opt, train_loader, val_loader, save_path)
Model Testing#
Test scores#
if not DO_TRAIN:
model = SmallCNN().to(DEVICE).to(torch.float32)
save_path = "./models"
model_url = "https://huggingface.co/AI4EIC/DNP2025-tutorial/resolve/main/FCALClassifier.safetensors"
model_path = download(model_url, save_path)
from safetensors import safe_open
tensors = {}
with safe_open(model_path, framework="pt", device="cpu") as f:
for key in f.keys():
tensors[key] = f.get_tensor(key)
model.load_state_dict(tensors)
⬇️ Downloading FCALClassifier.safetensors from https://huggingface.co/AI4EIC/DNP2025-tutorial/resolve/main/FCALClassifier.safetensors
FCALClassifier.safetensors: 98.3kB [00:00, 238kB/s]
✅ Download complete: ./models/FCALClassifier.safetensors
<All keys matched successfully>
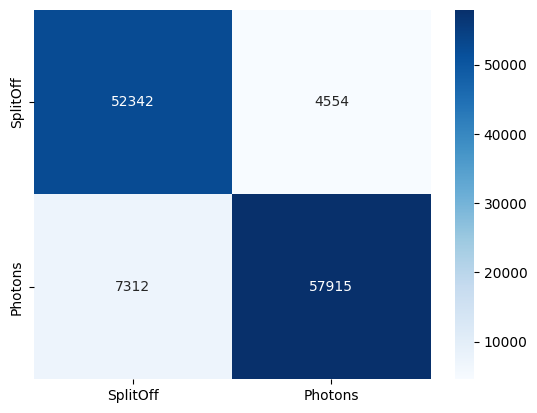
test_acc, test_auc, y_true, y_prob = evaluate(model, test_loader, desc="Test")
cm = confusion_matrix(y_true, (y_prob >= 0.5).astype(int), labels=[0, 1])
print(f"Test | acc {test_acc:.4f} | auc {test_auc:.4f}")
import seaborn as sns
import matplotlib.pyplot as plt
sns.heatmap(cm, annot=True, fmt="d", cmap="Blues", xticklabels=["SplitOff","Photons"], yticklabels=["SplitOff","Photons"])
plt.show()
Test | acc 0.9028 | auc 0.9626
# Reference values from https://arxiv.org/pdf/2002.09530
# 85% signal with 60% background rejection
import numpy as np
from sklearn.metrics import roc_curve, confusion_matrix
# y_true: 0 = photon (signal), 1 = split-off (background)
# y_prob: model score = P(split-off)
def eff_rej_at_threshold(y_true, y_prob, thr):
y_true = np.asarray(y_true, dtype=int)
y_prob = np.asarray(y_prob, dtype=float)
keep_photon = (y_prob < thr)
Nsig = np.sum(y_true == 0); Nbkg = np.sum(y_true == 1)
eps_sig = np.sum((y_true == 0) & keep_photon) / max(1, Nsig)
rej_bkg = np.sum((y_true == 1) & (~keep_photon)) / max(1, Nbkg) # predicted split-off
# Predicted label: 1=split-off, 0=photon (consistent with y_true)
y_pred = (y_prob >= thr).astype(int)
cm = confusion_matrix(y_true, y_pred, labels=[0, 1])
return eps_sig, rej_bkg, cm
def eff_rej_curve(y_true, y_prob):
# Standard ROC for positive class = split-off (1)
fpr, tpr, thrs = roc_curve(y_true, y_prob, pos_label=1)
# Map to your physics metrics:
eps_sig = 1.0 - fpr # photon efficiency
rej_bkg = tpr # split-off rejection
return eps_sig, rej_bkg, thrs
# --- Example usage ---
# If you have them already from evaluate():
# y_true, y_prob = <from your test set>
# 1) Curve over all thresholds
eps_sig, rej_bkg, thrs = eff_rej_curve(y_true, y_prob)
# 2) Pick a working point: match Barsotti–Shepherd ε_sig ≈ 0.85
target_eps = 0.85
i = int(np.argmin(np.abs(eps_sig - target_eps)))
thr_star = thrs[i]
eps_star, rej_star, cm_star = eff_rej_at_threshold(y_true, y_prob, thr_star)
print(f"Working point (ε_sig≈{target_eps:.2f}):")
print(f" threshold = {thr_star:.4f}")
print(f" ε_sig (photon eff) = {eps_star:.4f}")
print(f" R_bkg (split-off rej)= {rej_star:.4f}")
print(" Confusion matrix [rows=true, cols=pred] (0=photon,1=splitoff):")
print(cm_star)
# 3) If you just want your current threshold, e.g. t=0.5:
eps_05, rej_05, cm_05 = eff_rej_at_threshold(y_true, y_prob, 0.5)
print(f"\nAt t=0.50: ε_sig={eps_05:.4f}, R_bkg={rej_05:.4f}")
Working point (ε_sig≈0.85):
threshold = 0.2809
ε_sig (photon eff) = 0.8500
R_bkg (split-off rej)= 0.9380
Confusion matrix [rows=true, cols=pred] (0=photon,1=splitoff):
[[48362 8534]
[ 4044 61183]]
At t=0.50: ε_sig=0.9200, R_bkg=0.8879
Figure of Merit (FoM) Definitions#
In binary classifier optimization specifically one dealing with signal and backgrounds, the optimal threshold is chosen by maximizing a Figure of Merit (FoM) that quantifies the balance between signal efficiency and background suppression.
Signal Significance#
A commond way of defining signal significance is as follows,
where:
\( S = N_S \, \varepsilon_S \) is the number of selected signal events,
\( B = N_B \, \varepsilon_B \) is the number of selected background events,
\( \varepsilon_S \) is the signal efficiency (fraction of true signal retained),
\( \varepsilon_B \) is the background efficiency (fraction of background misclassified as signal).
This FoM estimates the expected statistical significance of signal observation in the presence of background.
Purity#
Purity measures how “clean” the selected sample is, i.e., the fraction of signal among all selected events:
\( \text{Purity} = \frac{S}{S + B} \)
Substituting in terms of efficiencies and total counts:
\( \text{Purity} = \frac{N_S \, \varepsilon_S}{N_S \, \varepsilon_S + N_B \, \varepsilon_B} \)
If the dataset’s signal-to-background ratio ( N_S / N_B ) is known, this becomes:
\( \text{Purity} = \frac{\varepsilon_S}{\varepsilon_S + \varepsilon_B / (N_S / N_B)} \)
Interpretation#
Signal Significance → best when you want to maximize discovery potential (e.g., physics searches).
Purity → best when you want clean signal selection (e.g., building templates or performing precise fits).
def find_optimal_threshold(y_true, y_prob, S_over_B=1.0):
"""
optimization using FoM maximization.
y_true: 0 = photon (signal), 1 = split-off (background)
y_prob: model score (probability of split-off)
S_over_B: expected signal-to-background ratio (S/B) in dataset
"""
fpr, tpr, thrs = roc_curve(y_true, y_prob, pos_label=1)
eps_sig = 1 - fpr
eps_bkg = 1 - tpr # background efficiency, not rejection
# Signal significance (TMVA default FoM)
# S/√(S+B)
fom = eps_sig / np.sqrt(eps_sig + eps_bkg / S_over_B + 1e-12)
purity = eps_sig / (eps_sig + eps_bkg / S_over_B + 1e-12)
i_opt = np.argmax(fom)
thr_opt = thrs[i_opt]
return {
"threshold": float(thr_opt),
"eps_sig": float(eps_sig[i_opt]),
"rej_bkg": float(1 - eps_bkg[i_opt]),
"SoverSqrtSB": float(fom[i_opt]),
"purity": purity[i_opt],
"curve": {"thresholds": thrs, "eps_sig": eps_sig, "rej_bkg": 1 - eps_bkg, "FoM": fom, "purity": purity},
}
# --- Example ---
opt = find_optimal_threshold(y_true, y_prob, S_over_B=1.0)
print(f"Optimal point Summary:")
print(f" threshold = {opt['threshold']:.3f}")
print(f" ε_sig = {opt['eps_sig']:.3f}")
print(f" R_bkg = {opt['rej_bkg']:.3f}")
print(f" FoM (S/sqrt(S+B)) = {opt['SoverSqrtSB']:.3f}")
print(f" Purity = {opt['purity']:.3f}")
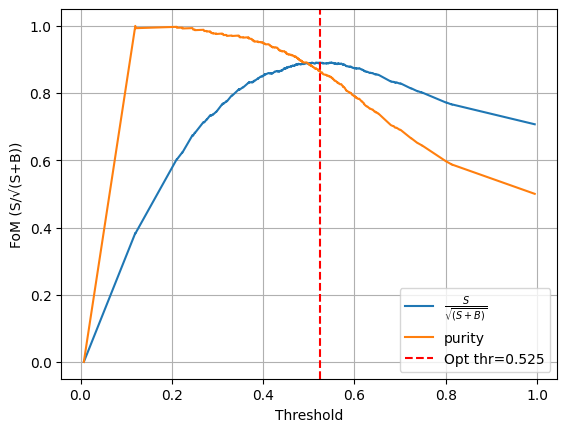
Optimal point Summary:
threshold = 0.525
ε_sig = 0.918
R_bkg = 0.857
FoM (S/sqrt(S+B)) = 0.891
Purity = 0.865
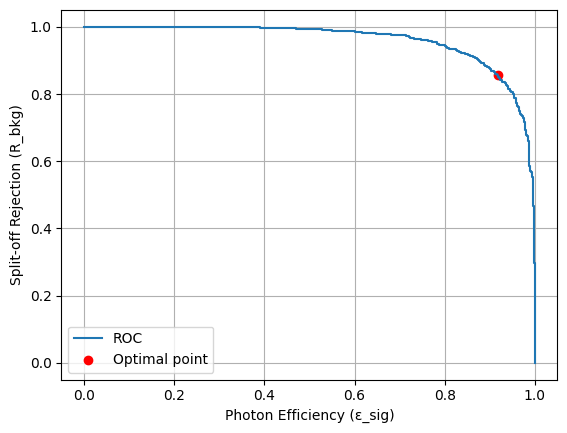
import matplotlib.pyplot as plt
plt.plot(opt["curve"]["eps_sig"], opt["curve"]["rej_bkg"], label="ROC")
plt.scatter(opt["eps_sig"], opt["rej_bkg"], color="r", label="Optimal point")
plt.xlabel("Photon Efficiency (ε_sig)")
plt.ylabel("Split-off Rejection (R_bkg)")
plt.legend()
plt.grid(True)
plt.show()
plt.figure()
plt.plot(opt["curve"]["thresholds"], opt["curve"]["FoM"], label = r"$\frac{S}{\sqrt{(S+B)}}$")
plt.plot(opt["curve"]["thresholds"], opt["curve"]["purity"], label = "purity")
plt.axvline(opt["threshold"], color="r", ls="--", label=f"Opt thr={opt['threshold']:.3f}")
plt.xlabel("Threshold")
plt.ylabel("FoM (S/√(S+B))")
plt.legend()
plt.grid(True)
plt.show()
Performance in bins of showerE#
EBins = [0.100, 0.500, 1.0, 2.0, 4.0]
test_acc, test_auc, y_true, y_prob, showerE = evaluate(model, test_loader, desc="Test", returnShowerE = True)
from sklearn.metrics import roc_auc_score, accuracy_score
def metrics_vs_energy(y_true, y_prob, showerE, EBins):
"""
Compute accuracy and AUC vs shower energy bins.
"""
EBins = np.asarray(EBins)
labels = [f"[{EBins[i]:.1f}, {EBins[i+1]:.1f})" for i in range(len(EBins)-1)]
results = []
for i in range(len(EBins)-1):
mask = (showerE >= EBins[i]) & (showerE < EBins[i+1])
if not np.any(mask):
results.append((labels[i], np.nan, np.nan, np.sum(mask)))
continue
yt = y_true[mask]
yp = y_prob[mask]
acc = accuracy_score(yt, yp >= 0.5)
auc = roc_auc_score(yt, yp) if np.unique(yt).size > 1 else np.nan
results.append((labels[i], acc, auc, np.sum(mask)))
return results
acc, auc, y_true, y_prob, showerE = evaluate(model, test_loader, returnShowerE=True)
EBins = [0.100, 0.500, 1.0, 2.0, 4.0]
bin_results = metrics_vs_energy(y_true, y_prob, showerE, EBins)
print(f"{'E_bin':<15}{'Acc':>10}{'AUC':>10}{'Nevents':>10}")
for label, acc_b, auc_b, n in bin_results:
print(f"{label:<15}{acc_b:10.3f}{auc_b:10.3f}{n:10d}")
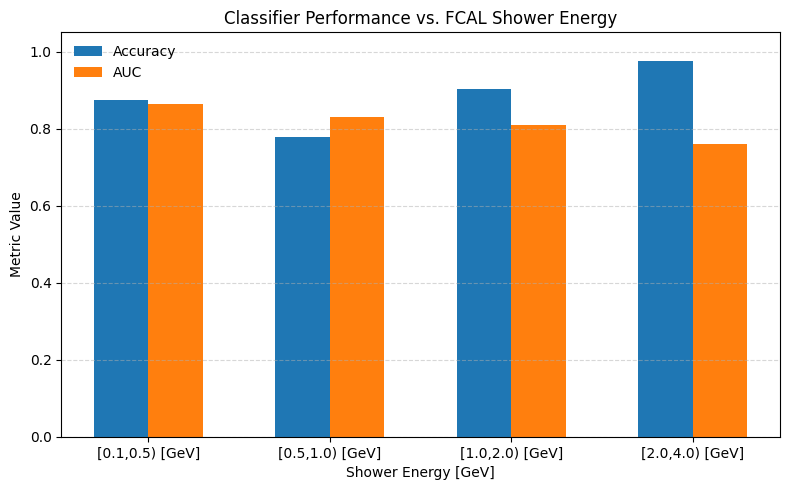
E_bin Acc AUC Nevents
[0.1, 0.5) 0.873 0.863 46603
[0.5, 1.0) 0.778 0.831 13455
[1.0, 2.0) 0.903 0.810 19009
[2.0, 4.0) 0.975 0.761 32519
import numpy as np
import matplotlib.pyplot as plt
centers = 0.5 * (np.arange(1, len(EBins)))
print (centers)
width = 0.15 # bar width
accs = [r[1] for r in bin_results]
aucs = [r[2] for r in bin_results]
plt.figure(figsize=(8,5))
# Horizontal offsets for each metric
plt.bar(centers - 1.5*width, accs, width, label="Accuracy")
plt.bar(centers - 0.5*width, aucs, width, label="AUC")
# Styling
plt.xlabel("Shower Energy [GeV]")
plt.ylabel("Metric Value")
plt.title("Classifier Performance vs. FCAL Shower Energy")
plt.xticks(centers - 0.15, [f"[{EBins[i]:.1f},{EBins[i+1]:.1f}) [GeV]" for i in range(len(EBins)-1)])
plt.ylim(0, 1.05)
plt.legend(frameon=False)
plt.grid(axis="y", linestyle="--", alpha=0.5)
plt.tight_layout()
plt.show()
[0.5 1. 1.5 2. ]
Inference on Physics events#
In order to extract the performance of CNN photon identifier. Let us use Physics events. We will be comparing the performance of CNN w.r.t rectangular cut and no cuts.
Mode |
Shower selection |
π⁰ combination rule |
Weights |
ω reconstruction |
|---|---|---|---|---|
CNN-based |
showers passing CNN photon score |
all photon pairs |
1/N (if multiple) |
combine π⁰ + π⁺π⁻ |
Rectangular cut |
showers within ±40 MeV of m(π⁰) |
all accepted |
1/N |
combine π⁰ + π⁺π⁻ |
Benchmark (no cut) |
all showers |
all possible |
none |
combine π⁰ + π⁺π⁻ |
Note: This is for pedagogical purposes only. A Physics based analysis would involve methods like Kinemattic Fitting which is beyond the scope of this tutorial
omega_url = "https://huggingface.co/datasets/AI4EIC/DNP2025-tutorial/resolve/main/formatted_dataset/CNN4FCAL_OMEGA_PATCHSIZE_11.h5"
omega_path = download("https://huggingface.co/datasets/AI4EIC/DNP2025-tutorial/resolve/main/formatted_dataset/CNN4FCAL_OMEGA_PATCHSIZE_11.h5", data_dir)
⬇️ Downloading CNN4FCAL_OMEGA_PATCHSIZE_11.h5 from https://huggingface.co/datasets/AI4EIC/DNP2025-tutorial/resolve/main/formatted_dataset/CNN4FCAL_OMEGA_PATCHSIZE_11.h5
CNN4FCAL_OMEGA_PATCHSIZE_11.h5: 400MB [00:08, 46.8MB/s]
✅ Download complete: data/CNN4FCAL_OMEGA_PATCHSIZE_11.h5
import h5py
with h5py.File(omega_path, "r") as f:
# Show top-level groups
eventKeys = list(f.keys())
print("Top-level keys:", eventKeys[:10])
print(f"\nContents of {eventKeys[0]}:")
for k, v in f[eventKeys[0]].items():
print(f" {k:20s} shape={v.shape} dtype={v.dtype}")
Top-level keys: ['event_000000', 'event_000001', 'event_000002', 'event_000003', 'event_000004', 'event_000005', 'event_000006', 'event_000007', 'event_000008', 'event_000009']
Contents of event_000000:
piMinus_p4 shape=(4,) dtype=float64
piPlus_p4 shape=(4,) dtype=float64
shower_p4 shape=(2, 4) dtype=float32
shower_patches shape=(2, 11, 11) dtype=float32
import numpy as np
import torch
PI0_MASS = 0.1349768 # GeV
OMEGA_MASS = 0.78265 # GeV
def fourvec_mass(p4):
"""Compute invariant mass from p4 = [px, py, pz, E]."""
px, py, pz, E = np.moveaxis(p4, -1, 0)
m2 = E**2 - (px**2 + py**2 + pz**2)
m2 = np.maximum(m2, 0.0)
return np.sqrt(m2)
def combine_p4(p4a, p4b):
"""Combine two 4-vectors."""
return p4a + p4b
def model_infer(model, patch, e0=ENERGY_SCALE, emax=CLIP_MAX):
"""Run CNN inference on one shower patch"""
model.eval()
patch_norm = log_global_norm(patch, e0=e0, emax=emax)
X = torch.tensor(patch_norm[None, None, :, :], dtype=torch.float32, device=DEVICE)
with torch.inference_mode():
probs = torch.softmax(model(X), dim=1)
pred_label = torch.argmax(probs).item() # 0 or 1
return pred_label # photon probability
from itertools import combinations
def analyze_event(ev, model, rect_mass_window=0.04, label: int=1):
"""
Run CNN, rectangular cut, and benchmark on one event.
Returns a dict with reconstructed pi0 and omega masses and weights.
"""
results = {"cnn": [], "rect": [], "bench": []}
piPlus = ev["piPlus_p4"]
piMinus = ev["piMinus_p4"]
showers = ev["shower_p4"]
patches = ev["shower_patches"]
# --- CNN inference on each shower
cnn_scores = [model_infer(model, patch) for patch in patches]
good_cnn = [i for i, s in enumerate(cnn_scores) if s == label] # threshold
#print (f"Good CNN : {good_cnn}")
# --- All combinations of showers for pi0
pairs = list(combinations(range(len(showers)), 2))
if not pairs:
return results # skip if < 2 showers
# --- 1. CNN-based
cnn_pairs = [p for p in pairs if p[0] in good_cnn and p[1] in good_cnn]
#print (f"Total Pairs : {len(pairs)}, Length of cnn_paris {len(cnn_pairs)}")
for pairs_sel, key in [(cnn_pairs, "cnn"), (pairs, "bench")]:
if not pairs_sel: continue
N = len(pairs_sel)
for (i, j) in pairs_sel:
p4_pi0 = combine_p4(showers[i], showers[j])
m_pi0 = fourvec_mass(p4_pi0)
p4_omega = combine_p4(p4_pi0, combine_p4(piPlus, piMinus))
m_omega = fourvec_mass(p4_omega)
results[key].append((m_pi0, m_omega, 1.0/N if key=="cnn" else 1.0))
# --- 2. Rectangular cut
rect_pairs = []
for (i, j) in pairs:
m_pi0 = fourvec_mass(combine_p4(showers[i], showers[j]))
if abs(m_pi0 - PI0_MASS) < rect_mass_window:
rect_pairs.append((i, j))
Nrect = len(rect_pairs)
for (i, j) in rect_pairs:
p4_pi0 = combine_p4(showers[i], showers[j])
m_pi0 = fourvec_mass(p4_pi0)
p4_omega = combine_p4(p4_pi0, combine_p4(piPlus, piMinus))
m_omega = fourvec_mass(p4_omega)
results["rect"].append((m_pi0, m_omega, 1.0/Nrect if Nrect>0 else 1.0))
return results
import h5py
from tqdm import tqdm
all_results = {"cnn": [], "rect": [], "bench": []}
PHOTON_LABEL = 1
PI0_MASS_CUT = 0.04 # fabs(pi0Mass - rec) <= 0.04 GeV
with h5py.File(omega_path, "r") as f:
ev_keys = [k for k in f.keys() if k.startswith("event_")]
for key in tqdm(ev_keys, desc="Running event inference"):
ev = {k: np.array(v) for k, v in f[key].items()}
out = analyze_event(ev, model, rect_mass_window=PI0_MASS_CUT, label=1)
for mode in all_results:
all_results[mode].extend(out[mode])
Running event inference: 100%|██████████| 97525/97525 [06:49<00:00, 237.96it/s]
import matplotlib.pyplot as plt
def plot_mass_spectrum(all_results, key="omega", nbins=80, mass_range=(0,1.2), ylog = False):
plt.figure(figsize=(7,5))
for label, color in zip(["bench", "rect", "cnn"], ["gray", "blue", "orange"]):
masses = [m[1 if key=="omega" else 0] for m in all_results[label]]
weights = [m[2] for m in all_results[label]]
plt.hist(masses, bins=nbins, range=mass_range, histtype="stepfilled",
alpha=0.4, label=label, weights=weights)
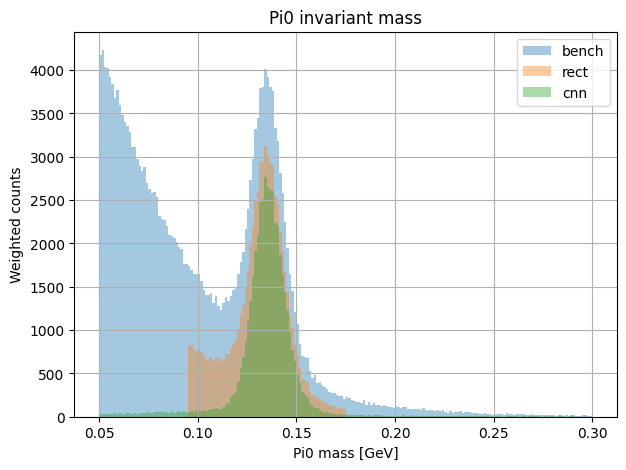
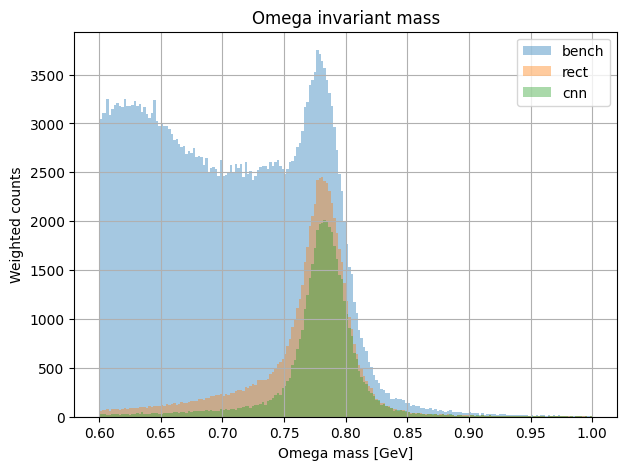
plt.xlabel(f"{key.capitalize()} mass [GeV]")
plt.ylabel("Weighted counts")
plt.title(f"{key.capitalize()} invariant mass")
plt.legend()
plt.grid(True)
plt.show()
plot_mass_spectrum(all_results, key="pi0", nbins = 200, mass_range=(0.05,0.3))
plot_mass_spectrum(all_results, key="omega", nbins = 200, mass_range=(0.6,1.0))
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
from scipy.special import wofz
# ------------------------
# Model definitions
# ------------------------
def voigt_plus_bkg(x, A, m0, gamma, sigma, b0, b1):
"""Voigtian (Lorentz ⊗ Gaussian) + linear background."""
z = ((x - m0) + 1j * gamma / 2) / (sigma * np.sqrt(2))
voigt = A * np.real(wofz(z)) / (sigma * np.sqrt(2 * np.pi))
return voigt + (b0 + b1 * x)
def voigt_only(x, A, m0, gamma, sigma):
"""Pure Voigtian signal component."""
z = ((x - m0) + 1j * gamma / 2) / (sigma * np.sqrt(2))
return A * np.real(wofz(z)) / (sigma * np.sqrt(2 * np.pi))
# ------------------------
# Fit utility
# ------------------------
def fit_mass_spectrum(masses, weights, label, color, range_omega=(0.6, 1.0), nbins=80):
"""Fit ω mass distribution with Voigt+linear background."""
hist, bins = np.histogram(masses, bins=nbins, range=range_omega, weights=weights)
centers = 0.5 * (bins[1:] + bins[:-1])
# Initial guesses
p0 = [max(hist), 0.782, 0.008, 0.02, np.min(hist), 0.0]
popt, pcov = curve_fit(voigt_plus_bkg, centers, hist, p0=p0, maxfev=10000)
A, m0, gamma, sigma, b0, b1 = popt
# Compute signal yield (area under Voigt)
signal_yield = np.trapz(voigt_only(centers, A, m0, gamma, sigma), centers)
# Plot
plt.plot(centers, voigt_plus_bkg(centers, *popt), color=color, lw=2, label=f"{label} fit")
plt.plot(centers, voigt_only(centers, A, m0, gamma, sigma), "--", color=color, alpha=0.6)
plt.plot(centers, b0 + b1*centers, "k--", alpha=0.3)
return {
"label": label,
"A": A,
"m0": m0,
"gamma": gamma,
"sigma": sigma,
"yield": signal_yield,
"cov": pcov,
"hist": hist,
"bins": bins,
"centers": centers
}
# ------------------------
# Run fits for CNN and rectangular selections
# ------------------------
plt.figure(figsize=(7,5))
# CNN selection
masses_cnn = [m[1] for m in all_results["cnn"]]
weights_cnn = [m[2] for m in all_results["cnn"]]
fit_cnn = fit_mass_spectrum(masses_cnn, weights_cnn, label="CNN", color="orange")
# Rectangular selection
masses_rect = [m[1] for m in all_results["rect"]]
weights_rect = [m[2] for m in all_results["rect"]]
fit_rect = fit_mass_spectrum(masses_rect, weights_rect, label="Rectangular", color="blue")
plt.bar(fit_cnn["centers"], fit_cnn["hist"], width=(fit_cnn["bins"][1]-fit_cnn["bins"][0]),
color="orange", alpha=0.3, label="CNN data")
plt.bar(fit_rect["centers"], fit_rect["hist"], width=(fit_rect["bins"][1]-fit_rect["bins"][0]),
color="blue", alpha=0.3, label="Rectangular data")
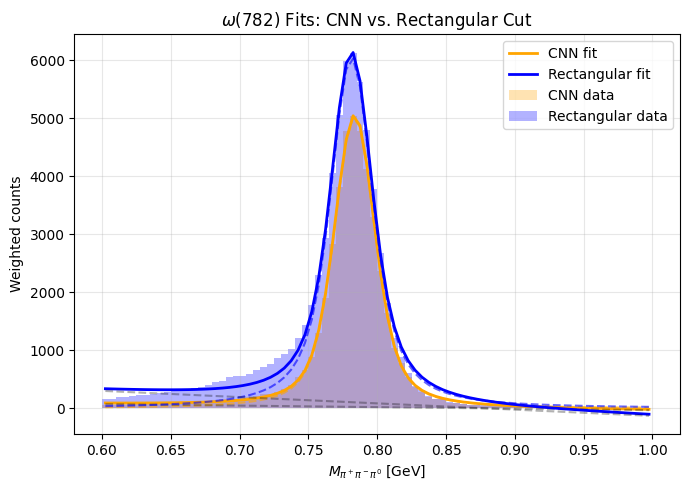
plt.xlabel(r"$M_{\pi^+\pi^-\pi^0}$ [GeV]")
plt.ylabel("Weighted counts")
plt.title(r"$\omega(782)$ Fits: CNN vs. Rectangular Cut")
plt.legend()
plt.grid(alpha=0.3)
plt.tight_layout()
plt.show()
# ------------------------
# Print summary
# ------------------------
print("=== Fit Comparison ===")
for f in [fit_rect, fit_cnn]:
print(f"\n[{f['label']}]")
print(f" m0 = {f['m0']:.5f} GeV")
print(f" gamma = {f['gamma']*1000:.2f} MeV (Lorentz width)")
print(f" sigma = {f['sigma']*1000:.2f} MeV (Gaussian smear)")
print(f" Yield = {f['yield']:.1f} (a.u.)")
/tmp/ipython-input-331058355.py:34: DeprecationWarning: `trapz` is deprecated. Use `trapezoid` instead, or one of the numerical integration functions in `scipy.integrate`.
signal_yield = np.trapz(voigt_only(centers, A, m0, gamma, sigma), centers)
=== Fit Comparison ===
[Rectangular]
m0 = 0.78137 GeV
gamma = 24.97 MeV (Lorentz width)
sigma = 8.79 MeV (Gaussian smear)
Yield = 299.6 (a.u.)
[CNN]
m0 = 0.78358 GeV
gamma = 17.15 MeV (Lorentz width)
sigma = 10.61 MeV (Gaussian smear)
Yield = 223.9 (a.u.)
Conclusion and Next steps#
From the preliminary CNN we built, We are able to get the following performance
Parameter |
Rectangular |
CNN |
Interpretation |
|---|---|---|---|
m₀ (GeV) |
0.78137 |
0.78358 |
Both close to PDG value (0.78265 GeV). |
Γ (MeV) |
24.97 |
17.15 |
CNN gives a ~30% narrower effective width, meaning reduced combinatorial contamination. |
σ (MeV) |
8.79 |
10.61 |
Comparable detector resolution; small variation due to shower phase space. |
Yield (a.u.) |
299.6 |
223.9 |
CNN yields fewer events but a cleaner signal. |
The CNN suppresses spurious photon–split-off combinations that broaden the ω peak. Even though it keeps fewer total events, the resulting spectrum is cleaner and closer to the intrinsic resonance shape — a clear indication that learned selection captures physical shower patterns better than rigid geometric cuts.
Excersises to try out#
Here are some concrete directions to extend this work and explore further physics–ML integration:
Optimize the classifier working point
Instead of using a hard
argmax, scan the photon probability threshold.Maximize a physics-driven metric such as \( S / \sqrt{S + B} \) or purity in ω reconstruction.
Compare how the yield–purity tradeoff affects the reconstructed mass width.
Model the background more realistically
Replace the linear or polynomial background with:
Chebyshev or Bernstein polynomials
Exponential or phase-space–motivated functions
Data-driven sideband templates
Study how background parameterization changes the extracted resonance parameters.
